So I get this question a lot.
How many replicates do I need for a proteomic profiling experiment?
 It's a great question and one unfortunately I have no idea how to answer with any accuracy.
It's a great question and one unfortunately I have no idea how to answer with any accuracy.I remember reading in a book somewhere (wish I remembered where) where the author stated that whenever he was asked this question he would say 30 biological replicates! Well that's great if you can actually generate 30 replicates and then pay for them to be analyzed....But alas in the world of proteomics, this is usually unobtainable.
So what to do....
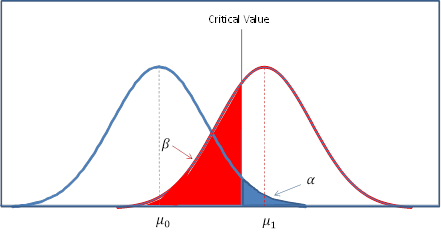
Let me start by explaining why I have no idea how to answer this question. It all has to do with Power and how it is calculated
Power is in essence your ability to detect a change in your sample when such a change actually exists. In other words what is your chance of committing a type II error (A false negative)
This is one of the best explanations of Power calculations I have seen (it's worth reading)
http://www.refsmmat.com/statistics/power.html
Some Bullet points
- Power depends on the number or replicates you do, your variation in your sample and the magnitude of the of the change you are trying to detect.
So the more replicates you analyze, the higher your chances of detecting a low magnitude change that has a large variation .
Now this can be calculated in a relatively straightforward way if you say have a small number of variables you are interested in and know your variation. I think anyway....
Now is that what we have in a proteomics or RNA-Seq experiment?
Unfortunately no
We have thousands of variables! And to make matters worse each one of those variables have their own variation and magnitude.
And it gets even more complicated with bottom up proteomics. Are you defining your variables as peptides or proteins? Ultimately we are interested in the the proteins (most of the time). But we need to reconstruct those proteins from the peptides we identify. Each of those peptides may have a different variation, (S/N) , and magnitude of change, across samples. How do we calculate power on a protein level....I have no idea.
For example
If you have a peptide that has a S/N of 1000:1 and a %CV of 5%, then you would need less replicates to detect a change in that peptide (depending on the magnitude of the change) than a peptide with a S/N of 2:1 with a %CV of 50%. Now lets say both of those peptides map back to the same protein....How do you calculate the power of that? This can actually be quite common as peptides can vary in their ionization by 4-5 orders of magnitude maybe (I need to look up he cite here) but I think that;s a reasonable guess and their ability to be cut by trypsin can vary by a lot as well (proteotypic vs non-proteotypic)
There has been a few papers on calculating power of proteomics experiments over the years, this one being one of the better ones.
If you look at this figure From the above paper
You can see a larger version of this figure in the paper, but essentially if you can somehow know that your protein has a 50% variation between groups (how you calculate this I have no idea...) at a power of 0.8 (one in five chance of detecting it) you would need 15 biological replicates per group to detect a 1.5 fold change. If you have a 25% variation you would only need 5 replicates. The good news is if you have a 100% variation you may need only 5 replicates to detect a 3 fold change. I think that's probably realistic...
Does the protein you are interested in have a 50% variation, 25% variation, 5% variation? I really have no idea. Most likely in your 1000+ plus proteins we can identify you will have some with a low variation (usually the high abundant and less interesting ones) and some with a large variation (less abundant more interesting ones) .
One can only get a handle on their variation after the experiment is run. You can begin to calculate your %CV's and S/N (1/CV) but I don't know how to do this before the experiment is run.
So more replicates the better, less variation the better. But it's still possible that you will need that 30+ replicates to see that < 1 fold change.....



